Restoring the history of texts: ordinary and genetic. "People and their genes"
Report at the Round Table: “Genetics - a bridge between the natural and human sciences” of the V Congress of the Vavilov Society of Genetics and Breeders (Moscow, 06/26/2009)
The topic of our report: studying human migrations using genetic data - both in historical and prehistoric times.

And the theme of the entire “Round Table” is a technical examination of the bridge that genetics is building across the gap between the humanities and natural sciences.

Genogeography is no longer a young science, and therefore it has been building this bridge for more than eighty years. The founder of genogeography, Alexander Sergeevich Serebrovsky, insisted that genogeography is a historical science, not a biological one. He believed that genogeography, using genetic markers, should describe the history of populations and human migration routes. A.S. himself Serebrovsky used the phenotypes of Dagestan chickens as a genetic marker - differences between chicken populations indicated differences between the gene pools of their hosts, the intensity of gene exchange (and exchange of chickens) between different gorges of Dagestan. Here is a diagram of such a study. Suppose that in one gorge there are only red chickens, in another there are black chickens, in a third there are only white chickens.
 |
New powerful markers of population history have appeared in the arsenal of genetics - "uniparental" markers. Mitochondrial DNA (mtDNA), transmitted through generations along the maternal line, was the first to gain popularity: it made it possible to convincingly substantiate the monocentric theory of the origin of humanity and the “exit from Africa” as the most important stage in the settlement of modern humans across the planet. At the height of the boom in mtDNA research, when most population geneticists focused on its study, another genetic system quickly entered the scene - the Y chromosome, inherited through generations along the paternal line. Although it has not yet succeeded in displacing mtDNA as a leader, the Y chromosome has confidently taken its place next to it. The resulting duet has become a generally accepted standard in world research. What is the appeal of these markers? The absence of recombination makes it possible to reconstruct the chain of successively occurring mutations (from Adam or from Eve), to determine the place and time of their occurrence, and, consequently, to trace the process of human settlement on the planet.
Therefore, modern genogeography can be called the science of typos. If there were no typos - mutations - in genetic texts, then genogeography would have nothing to study: all men would have identical Y chromosomes, and women would have identical copies of the same mtDNA molecule. Mutations serve as the same markers as the errors of chronicle copyists - thanks to their errors, it is possible to give a relative dating of different editions of the chronicles: those editions that included both old “typos” and their own are considered as later.
Based on genetic typos, you can build phylogenetic tree the origin of all modern genetic lines from one original one and reveal the most ancient genetic relationship of the population of different continents. The most ancient mutations will determine the main, largest branches of the tree of the Y chromosome or mtDNA ( haplogroups). Later mutations show how these branches branch into smaller ones ( subhaplogroups). Lots of leaves ( haplotypes) differ only in the most recent mutations and cover the entire tree, reflecting the genetic diversity of modern humanity.
 |
If we superimpose the frequencies of occurrence of various mutations on geographical map, then we will see areas of their accumulation - those regions in which, by the will of history, these typos have multiplied. The longer a population developed in this region, the more mutations it could accumulate. Its daughter populations took with them only a small part of this diversity when they set off. Therefore, we can also detect those daughter regions into which waves of migrations brought certain haplogroups and haplotypes. And knowing the relative timing of mutations will help separate ancient migrations from more recent ones.
 |
So, if we look at the slide as well Where are each of these schematic haplotypes geographically distributed? We see that the most ancient ones are widespread in Africa (everyone has the African “red” mutation), and then the right branch goes to Asia (all haplotypes have the “blue” Asian mutation), and the left branch (with European “green” mutations) to Europe . That is, we have restored the picture of the most important migration in the history of mankind - the picture of the exit from Africa.
Of course, these are only the very basics, the “skeleton” of the tool that genogeography uses to track ancient and historical migrations. It is easier to understand the capabilities and limitations of this tool using live examples of genogeographic work.
 |
Of course, it is impossible to talk about all the variety of genetic studies studying population migrations. Therefore, we limited ourselves to only those works in which we ourselves participated in collaboration with many other colleagues. We have imposed one more restriction - the work must be fresh - completed within the last two years. The resulting set of works is shown on the slide. They cover vast times and spaces: in terms of dates, the extreme points differ by a thousand times (from 140,000 years to 140 years), and in terms of geography, they cover the space from South Africa to the Russian North and the Pamirs.
Such a selection of studies from world science will be almost random - and, since we did not select the works, it will outline to you not only the advantages, but also the possible disadvantages of what is being built bridge between the humanities and natural sciences.
 |
SOUTH AFRICA: AT THE DAWN OF MODERN HUMANITY.
The first study we report outlines the African portion of the global mtDNA family tree. Analysis of complete nucleotide sequences was carried out in South African populations mitochondrial DNA. This labor-intensive work was necessary to answer the question - what were the very first stages of microevolution of Homo sapiens? The main result of this work was the clarification of the phylogenetic tree of humanity. Let us indicate two most important features.
Firstly, mtDNA claims that 140,000 years ago the tree split into two large trunks - the Khoisan - and the rest of humanity. The abstracts of the next report (Dybo, Starostin, 2009) say that linguists also contrast the Khoisan languages with the languages of the rest of humanity. So a piece of the bridge between humanists and geneticists has come to light.
The second feature is already known from earlier works, but no less surprising. This tree also shows that all genetic diversity is concentrated in Africa, and the haplogroups of all other continents are just two skinny branches on the African trunk (shown in pink). We see that very few Africans left their homeland to populate the rest of the world - Eurasia, America, Australia. This tree illustrates well general principle tracking migrations - dispersing populations, breaking away from the original massif, take with them on their journey only a small part of the branches, a small part of the available genetic diversity. Further microevolution leads to the growth of new secondary subhaplogroups in different regions of the planet, allowing us to trace more and more recent migrations.
 |
SOUTH AFRICA: GIANTS AND Dwarfs.
Let's skip half the time scale and find ourselves in Central Africa at around 70,000 years ago. When Louis Quintano-Murchi asked for access to our database for comparative analysis, I was very happy, because in my early youth I read Nikolai Gumilyov’s stories about these equatorial forests: “I pitched a tent on a stone slope of the Abyssinian mountains running down to the west, And carelessly watched the sunsets blaze, Above the green roof of distant forests.”. But then a dying Frenchman came out of these mysterious forests to Gumilev, telling about the death of their expedition in the country of cannibal pygmies.
Fortunately, the expedition of our French colleagues was more successful, and we studied the gene pools of the shortest and tallest populations of the planet - the pygmies and Bantu-speaking peoples of Africa. mtDNA states that 70 thousand years ago they were still a single population. Their separation was caused by the climate crisis in the history of our planet. Ice ages in the history of the earth had no less catastrophic consequences for Africa than for Europe. It was a time of drying out of the planet - forests disappeared, their place was taken by savannas and deserts. Arose ecological boundary, which divided the ancestors of the Pygmies and the Bantu. Many thousands of years passed, and both populations acquired peculiar anthropological features. When their ranges overlapped again, the flow of genes between them, as mtDNA showed, became one-way: only Bantu men married small pygmy women, who brought their mtDNA haplogroups. Reverse gene flow has not been detected - the mtDNA lines of the Bantu-speaking peoples of Africa are not traced among the pygmies.
 |
Neolithic Europe: paleoDNA of ancient populations.
The first wave of European settlement is associated with the Paleolithic. Second wave - Mesolithic recolonization Europe after the glacier retreat. But the third wave is the most controversial - Neolithic farmers(the slide on the left shows mathematical modeling of the spread of agriculture in Europe).
In the classic work of archaeologist Ammermann and geneticist Cavalli-Sforza, a hypothesis was formulated "demic spread": it was the third – Neolithic – wave of settlement of farmers that formed the main features of the European gene pool. However, mtDNA data subsequently indicated a Paleolithic age for most European haplogroups. This became the basis for an alternative hypothesis "cultural diffusion": migration of agriculture without farmers. Both of these approaches reconstructed the gene pools of past eras from the genetic structure of their modern descendant populations.
But only ancient DNA data (obtained in reliable laboratories and received worldwide recognition) provide direct information about the gene pool of ancient populations. A study of paleoDNA from one of the first Neolithic cultures in Europe - Linear Band Pottery (red oval on the map on the left) - unexpectedly revealed a high frequency of mtDNA haplogroup N1a, which is almost never found among modern Europeans. This may mean that the first agricultural population of Europe actually left almost no descendants. New data obtained by the same group of researchers in collaboration with our team made it possible to clarify this conclusion: they discovered the Middle Eastern roots of the first farmers in Europe. Their migration proceeded approximately as shown by the red arrows. But most modern Europeans have a completely different gene pool. This means that the emergence of agriculture in Europe was associated with the migration of the first farmers, which was not numerous, and the subsequent spreading agriculture within Europe was mainly "cultural borrowing".
Although this is a kind of compromise between the “demic” and “cultural” hypotheses of the spread of agriculture: spreading agriculture within Europe had the character of “cultural diffusion”, but the emergence of agriculture in Europe is associated with the distant migration of the first farmers.
 After a couple of thousand years, the time came for the return migration - from Europe to the Middle East. We're talking about the Crusades. As you know, at the call of the pope, knights from most Western European states went to Palestine, where their states existed for more than a hundred years. The question of the genetic consequences of these events remained open - from historical data it is difficult to understand how many European settlers remained in the Levant. But genogeography has revealed a specific haplotype (red circle) in the modern population of Lebanon. As you can see, in the east this haplotype is not found anywhere else (there are only blue circles around: the absence of this haplotype). But it is in the west (red circles), and its geography even repeats the geography of the countries participating in the crusades: this haplotype is found in the gene pools of all participating countries (and, of course, outside them - this is the “European” haplotype). This was an example of a period for which there are already written sources. But even for historically reliable migrations, the question remains: was this event only historical or did it leave a trace in genetics? There are also events unknown to written history. Here genetics can reveal unexpected facts. After a couple of thousand years, the time came for the return migration - from Europe to the Middle East. We're talking about the Crusades. As you know, at the call of the pope, knights from most Western European states went to Palestine, where their states existed for more than a hundred years. The question of the genetic consequences of these events remained open - from historical data it is difficult to understand how many European settlers remained in the Levant. But genogeography has revealed a specific haplotype (red circle) in the modern population of Lebanon. As you can see, in the east this haplotype is not found anywhere else (there are only blue circles around: the absence of this haplotype). But it is in the west (red circles), and its geography even repeats the geography of the countries participating in the crusades: this haplotype is found in the gene pools of all participating countries (and, of course, outside them - this is the “European” haplotype). This was an example of a period for which there are already written sources. But even for historically reliable migrations, the question remains: was this event only historical or did it leave a trace in genetics? There are also events unknown to written history. Here genetics can reveal unexpected facts. |
 Another event, covered in detail by written history, but around which there is heated debate. Tatar-Mongol yoke Some call it a grave catastrophe for the Eastern Slavs, while Eurasians consider it a happy occasion for the birth of Russian statehood. These questions are not related to genetics, but one can often hear the opinion that the Russian gene pool has become intermediate between the peoples of Europe and Central Asia. And here the word belongs to genetics. Another event, covered in detail by written history, but around which there is heated debate. Tatar-Mongol yoke Some call it a grave catastrophe for the Eastern Slavs, while Eurasians consider it a happy occasion for the birth of Russian statehood. These questions are not related to genetics, but one can often hear the opinion that the Russian gene pool has become intermediate between the peoples of Europe and Central Asia. And here the word belongs to genetics. Genetic traces of aliens from the east cannot be detected. This mtDNA genetic distance map shows the purely European roots of the Russian gene pool (blue tones) and the foreignness of the gene pools of Central Asia (brown tones). And the analysis of all other markers leads to the same conclusions - from the Y chromosome to the study of the dental system. |
 What about the return migration, when several centuries later the Russians began to conquer Asia? Genetic differences between the indigenous population of the Caucasus (major haplogroups G and J are indicated in blue) and Eastern Slavs(major haplogroups R1a and I are indicated in red) are very clear. We studied two groups of Cossacks in the North Caucasus. It turned out that the Kuban Cossacks are genetically indistinguishable from Russians and Ukrainians. And the Terek Cossacks absorbed almost half of the local Caucasian haplotypes (Blue colour). This is also an example of when genetics introduces new information even for those historical events that are considered well documented. What about the return migration, when several centuries later the Russians began to conquer Asia? Genetic differences between the indigenous population of the Caucasus (major haplogroups G and J are indicated in blue) and Eastern Slavs(major haplogroups R1a and I are indicated in red) are very clear. We studied two groups of Cossacks in the North Caucasus. It turned out that the Kuban Cossacks are genetically indistinguishable from Russians and Ukrainians. And the Terek Cossacks absorbed almost half of the local Caucasian haplotypes (Blue colour). This is also an example of when genetics introduces new information even for those historical events that are considered well documented. |
 Surnames are a hallmark of linguistics, and their use to study gene pools is a clear bridge between the two sciences. There are four ways to combine surnames with genetics, but we will only talk about the fourth, which arose in Russia over Last year thanks to the interest of our fellow citizens in their surnames. This RGNF project “Same namesakes or relatives?”. For groups of namesakes, we provide free analysis of their Y chromosomes. If they are identical, then people received both the surname and the Y chromosome from one common ancestor, which means they are relatives. If the Y chromosomes are different, they are only namesakes. Surnames are a hallmark of linguistics, and their use to study gene pools is a clear bridge between the two sciences. There are four ways to combine surnames with genetics, but we will only talk about the fourth, which arose in Russia over Last year thanks to the interest of our fellow citizens in their surnames. This RGNF project “Same namesakes or relatives?”. For groups of namesakes, we provide free analysis of their Y chromosomes. If they are identical, then people received both the surname and the Y chromosome from one common ancestor, which means they are relatives. If the Y chromosomes are different, they are only namesakes. At the moment, about four hundred people representing sixty families have been analyzed. This picture from our website shows that, for example, two participants shown in dark green are related to each other - they differ in only one microsatellite out of seventeen STR markers, and the other participant (light green) differs from them in two others STR markers. |
 Let's show it with one example. Of all the continents of the world, the gene pool of Europe has been studied in more detail. And in Europe the simplest and most well-documented history is Icelandic gene pool. A thousand years ago, this uninhabited island was colonized by Vikings from Scandinavia. But they also brought slaves from the British Isles. The question is: in what proportion are these gene pools combined?. The simplest question, the most studied region, but every new genetic study provides a new answer. Links to 6 works are given. Their results: from Britain's share of 98% to Scandinavia's share of 80%. And imagine what a humanities specialist must think after reading these studies. Will he believe even one more conclusion made by geneticists? According to our observations, they still believe. But the most insightful are already moving from trust to skepticism. Let's show it with one example. Of all the continents of the world, the gene pool of Europe has been studied in more detail. And in Europe the simplest and most well-documented history is Icelandic gene pool. A thousand years ago, this uninhabited island was colonized by Vikings from Scandinavia. But they also brought slaves from the British Isles. The question is: in what proportion are these gene pools combined?. The simplest question, the most studied region, but every new genetic study provides a new answer. Links to 6 works are given. Their results: from Britain's share of 98% to Scandinavia's share of 80%. And imagine what a humanities specialist must think after reading these studies. Will he believe even one more conclusion made by geneticists? According to our observations, they still believe. But the most insightful are already moving from trust to skepticism. |
 |
Therefore, reconstruction of the bridge is necessary - and this is the third part of our report.
 |
The fifth pillar - and we consider it one of the main ones - participation of geneticists and humanists in joint projects. In the last month alone I have participated in three - in America, Spain and Russia.
The Genography project includes such venerable specialists as the archaeologist Lord Renfew, the author of the classification of the world's languages, Merritt Ruhlen, and Miiv Leakey from the dynasty of paleoanthropologists. Their timely consultations sometimes save us from... inaccuracies.
In other projects, communication with humanities specialists grows to genuine cooperation. This is a project for the initial settlement of the Arctic and Subarctic and a project for the neolithization of Europe.
The second meeting took place in Spain. The three-year project aims to model the Neolithic settlement of Europe. The working group led by Pavel Markovich Dolukhanov included mainly mathematicians, archaeologists, paleogeographers and geneticists. A volume of the team's works has already been published.
The third project is in Russia. His task is human settlement of the North of Eurasia. The working group included paleogeographers, paleozoologists, paleobotanists, geneticists, anthropologists, daters and many archaeologists from all regions of the country. The result of the work will be a collective monograph-Atlas.
 |
Finally, a purely genetic support that helps strengthen the reliability of the conclusions is multisystem approach. For example, having discovered similarities in the variability of anthropological characteristics, classical and DNA markers, there is no doubt about the objectivity of the longitudinal pattern. We wrote a whole book about this approach (see monograph "Russian gene pool on the Russian plain"), but we won’t have time to tell it all here.
An important step along this path is the simultaneous use of data on mtDNA and the Y chromosome: in this case, only those results that are confirmed by both systems should be considered reliable.
However, both of these systems are essentially very similar: both are haploid, both do not recombine, both are analyzed by the same phylogeographic methods, both are most vulnerable to the effects of genetic drift. And this can lead to distortions in the reconstructed migration picture.
Therefore the next step is testimonies of many eyewitnesses, that is, expanding the range of analyzed genetic systems due to autosomal DNA and classical gene markers, as well as the inclusion of informative quasi-genetic systems - surnames, anthropological, archaeological and linguistic characteristics. When pictures of the world - Russian, European, Eurasian - coincide despite the fact that they are depicted by completely different witnesses (genetics, anthroponymics, anthropology), we can be sure that the genetic traces of migrations are real and reliable.
Using many systems – multisystem approach– opens the way to a real synthesis of knowledge about the history of human populations obtained by different sciences themselves.
 |
We hope that thanks to these and other supports, the genetic bridge will become not only fashionable, but also a reliable meeting place for representatives of the natural sciences and humanities.
Laboratory of Population Genetics, State University of Moscow State Scientific Center, Russian Academy of Medical Sciences
Genofond.ru
Where did the Russians come from? Who was our ancestor? What do Russians and Ukrainians have in common? For a long time, the answers to these questions could only be speculative. Until geneticists got down to business.
Adam and Eve
Population genetics deals with the study of roots. It is based on indicators of heredity and variability. Geneticists have discovered that all of modern humanity can be traced back to one woman, whom scientists call Mitochondrial Eve. She lived in Africa more than 200 thousand years ago.
We all have the same mitochondrion in our genome - a set of 25 genes. It is transmitted only through the maternal line.
At the same time, the Y chromosome in all modern men is also traced back to one man, nicknamed Adam, in honor of the biblical first man. It is clear that we are only talking about the closest common ancestors of all living people; their genes came to us as a result of genetic drift. It is worth noting that they lived at different times - Adam, from whom all modern males received their Y chromosome, was 150 thousand years younger than Eve.
Of course, it’s a stretch to call these people our “ancestors,” since out of the thirty thousand genes that a person possesses, we have only 25 genes and a Y chromosome from them. The population increased, the rest of the people mixed with the genes of their contemporaries, changed, mutated during migrations and the conditions in which people lived. As a result, we received different genomes of different peoples that subsequently formed.
Haplogroups

It is thanks to genetic mutations that we can determine the process of human settlement, as well as genetic haplogroups (communities of people with similar haplotypes who have a common ancestor who had the same mutation in both haplotypes) characteristic of a particular nation.
Each nation has its own set of haplogroups, which are sometimes similar. Thanks to this, we can determine whose blood flows into us and who our closest genetic relatives are.
According to a 2008 study conducted by Russian and Estonian geneticists, the Russian ethnic group genetically consists of two main parts: the inhabitants of Southern and Central Russia are closer to other peoples who speak Slavic languages, and the indigenous northerners are closer to the Finno-Ugrians. Of course, we are talking about representatives of the Russian people. Surprisingly, there is practically no gene inherent in Asians, including the Mongol-Tatars. So the famous saying: “Scratch a Russian, you will find a Tatar” is fundamentally wrong. Moreover, the Asian gene also did not particularly affect the Tatar people; the gene pool of modern Tatars turned out to be mostly European.
In general, based on the results of the study, in the blood of the Russian people there is practically no admixture from Asia, from the Urals, but within Europe our ancestors experienced numerous genetic influences from their neighbors, be they Poles, Finno-Ugric peoples, peoples of the North Caucasus or ethnic group Tatars (not Mongols). By the way, haplogroup R1a, characteristic of the Slavs, according to some versions, was born thousands of years ago and was common among the ancestors of the Scythians. Some of these Proto-Scythians lived in Central Asia, while others migrated to the Black Sea region. From there these genes reached the Slavs.
Ancestral home

Once upon a time, Slavic peoples lived on the same territory. From there they scattered around the world, fighting and mixing with their indigenous population. Therefore, the population of current states, which are based on the Slavic ethnic group, differ not only in cultural and linguistic characteristics, but also genetically. The further they are geographically from each other, the greater the differences. Thus, the Western Slavs found common genes with the Celtic population (haplogroup R1b), the Balkans with the Greeks (haplogroup I2) and the ancient Thracians (I2a2), and the Eastern Slavs with the Balts and Finno-Ugrians (haplogroup N). Moreover, interethnic contact of the latter occurred at the expense of Slavic men who married aboriginal women.
Despite the many differences and heterogeneity of the gene pool, Russians, Ukrainians, Poles and Belarusians clearly fit into one group on the so-called MDS diagram, which reflects genetic distance. Of all the nations, we are closest to each other.
Genetic analysis makes it possible to find the above-mentioned “ancestral home where it all began.” This is possible due to the fact that each migration of tribes is accompanied by genetic mutations, which increasingly distort the original set of genes. So, based on genetic proximity, the original territorial one can be determined.
For example, according to their genome, Poles are closer to Ukrainians than to Russians. Russians are close to southern Belarusians and eastern Ukrainians, but far from Slovaks and Poles. And so on. This allowed scientists to conclude that the original territory of the Slavs was approximately in the middle of the current settlement area of their descendants. Conventionally, the territory of the subsequently formed Kievan Rus. Archaeologically, this is confirmed by the development of the Prague-Korchak archaeological culture of the 5th-6th centuries. From there the southern, western and northern waves of Slavic settlement had already begun.
Genetics and mentality

It would seem that since the gene pool is known, it is easy to understand where the national mentality comes from. Not really. According to Oleg Balanovsky, an employee of the Laboratory of Population Genetics of the Russian Academy of Medical Sciences, between national character and there is no connection with the gene pool. These are already “historical circumstances” and cultural influences.
Roughly speaking, if a newborn baby from a Russian village with a Slavic gene pool is taken directly to China and raised in Chinese customs, culturally he will be a typical Chinese. But as far as appearance and immunity to local diseases are concerned, everything will remain Slavic.
DNA genealogy

Along with population genealogy, today private directions for studying the genome of peoples and their origins are emerging and developing. Some of them are classified as pseudo-sciences. For example, Russian-American biochemist Anatoly Klesov invented the so-called DNA genealogy, which, according to its creator, “is a practically historical science, created on the basis of the mathematical apparatus of chemical and biological kinetics.” Simply put, this new direction is trying to study the history and time frame of the existence of certain clans and tribes based on mutations in male Y chromosomes.
The main postulates of DNA genealogy were: the hypothesis of the non-African origin of Homo sapiens (which contradicts the conclusions of population genetics), criticism of the Norman theory, as well as the extension of the history of the Slavic tribes, which Anatoly Klesov considers the descendants of the ancient Aryans.
Where are such conclusions from? Everything is from the already mentioned haplogroup R1A, which is the most common among the Slavs.
Naturally, such an approach gave rise to a sea of criticism, both from historians and geneticists. In historical science, it is not customary to talk about the Aryan Slavs, since material culture (the main source in this matter) does not allow us to determine continuity Slavic culture from the peoples of Ancient India and Iran. Geneticists even object to the association of haplogroups with ethnic characteristics.
Doctor of Historical Sciences Lev Klein emphasizes that “Haplogroups are not peoples or languages, and giving them ethnic nicknames is a dangerous and undignified game. No matter what patriotic intentions and exclamations it hides behind.” According to Klein, Anatoly Klesov's conclusions about the Aryan Slavs made him an outcast in the scientific world. How the discussion around Klesov’s newly announced science and the question of the ancient origins of the Slavs will further develop remains anyone’s guess.
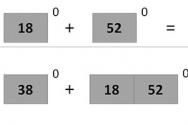
0,1%

Despite the fact that the DNA of all people and nations is different and in nature there is not a single person identical to another, from a genetic point of view we are all extremely similar. All the differences in our genes that gave us different colour skin and eye shape, according to Russian geneticist Lev Zhitovsky, make up only 0.1% of our DNA. For the remaining 99.9% we are genetically the same. Paradoxical as it may seem, if we compare various representatives human races and our closest relatives, chimpanzees, then it turns out that all people differ much less than chimpanzees in one herd. So, to some extent, we are all one big genetic family.
Once upon a time, anthropologists around the world relied only on a spade and a spade in their quest to unearth chronological evidence of the development of great apes, the emergence of Homo sapiens and the colonization of the entire planet by this species.
In the 21st century, archaeological finds were replaced by data from a global genographic project, which simultaneously revealed to the whole world the significant genetic diversity of all humanity and outlined the sequence of human settlement on Earth. For example, conclusions based on excavations of ancient settlements of American Indians were confirmed by geneticists, and now everyone knows for certain that the Indians are the descendants of indigenous Siberians who moved to the coast of North America more than 15 thousand years ago.
At the same time, attempts to conduct a more detailed analysis of successive waves of human migration across the planet based only on genetic data have so far been difficult. And often anthropologists try to resort to genetic analysis only to confirm controversial anthropological theories, which need to be substantiated traditional methods fails.
As a result, all the conclusions of genetic anthropology are covered with a certain layer of doubt - especially since they often turn out to be quite unexpected. However, the huge amount of information hidden in human chromosomes would be foolish not to use, and we are already seeing how genetic methods are increasingly used in the study of the history of human development.
The history of human migration may soon acquire a lot of new details thanks to a new method for analyzing genetic diversity.
developed by specialists from British Oxford and American Cornell University under the leadership of Daniel Falus from the University of the Irish city of Cork. The new technique is especially useful for comparing the collective genomes of entire human populations and individual samples, which allows us to say much more about genetic relatedness than previously used techniques.
The new statistical approach promises to establish the time frame of at least the main migration forks in human settlement, as well as the sizes of populations that separated at these forks and reunited later in modern peoples. Their results scientists published in PLoS Genetics.
It has three main advantages over currently used techniques.
The first of them is the adaptability of the method to take into account block copying of entire DNA sections when mixing human populations. Mutations during inheritance occur in two main ways: point mutations, when individual “letters” of the genetic code are subject to change; and block, when long sections of DNA are copied, rearranged or disappeared. The methods used so far have only taken into account point mutations, without being able to adequately examine the complex interdependence between different elements of the genome. Geneticists themselves sometimes call such methods “plush” genetics behind their backs.
Thus, in essence, the model includes not only existing populations found in real data, but also synthetic ancestral populations.
Of course, in reality, it is not real ancestral peoples that are modeled, but their surrogates, built from real data based on the hierarchy of populations assumed for each specific scenario, sorted in the descendant-ancestor relation.
As the authors note, such a model should work especially well in the case of sequential settlement of people region after region, since in this case a smaller number of hypothetical populations has to be “completed”, and the matter only comes down to choosing the correct sequence of “seniority” of peoples. According to modern ideas, this is approximately how the settlement of people happened: we all came out of Africa and then colonized more and more new territories.
Scientists tested their model on data from the Human Genome Diversity Project (HGDP), published in 2006.
They include genetic information on almost a thousand representatives of 53 various peoples. For each DNA donor, the data contains information on 2 thousand genetic markers; Although the method of Felush and his colleagues is adapted for processing large amounts of data on copying individual sections of the genome, it can also work with relatively “sparse” data, characteristic of the so far traditional approach.
Based on these data, scientists have identified nine main stages of colonization of our planet. Geneticists note that these stages did not necessarily occur in a clear chronological order, but are confident that they have correctly identified the main ancestors of each population at each stage.
It is not surprising that the settlement of the world by humans occurs first in Africa - from the communities of hunters and gatherers in southern Africa to the center and north of the continent and then through the Middle East to central part Eurasia. The peoples living here, including the Russian Adygeis, are strongly mixed with each other, which, according to the authors, indicates the absence of any noticeable “bottlenecks” in the settlement of people during this period.
Among the European peoples, the model identifies the French, Italians and Tuscans as “progenitors”, in whose genes a strong signal was found from the indigenous inhabitants of central Africa. At the same time, populations from the HGDP living on the fringes of Europe - Sardinians, Basques, Orkney Islanders and Russians - received large amounts of genetic material from both Europeans and the Middle East and the center of Eurasia, absorbing newcomers to Europe during waves of migration following the main one. Russians, by which we mean the inhabitants of the north of the European part of Russia, have especially many ancestors from various regions of Eurasia.
The analysis also revealed some very unexpected details.
For example, the Yakuts had ancestors not only among the Russians, which is unlikely to surprise anyone, but also among the inhabitants of the Orkney Islands, located north of Scotland.
In addition, the inhabitants of South America borrowed a certain number of genes from the Mongols, while the North American Indians descended for the most part from the peoples who now inhabit the more northern regions of Siberia and do not have Mongolian roots. However, the blood of Siberians in the veins of South Americans remains dominant. All this may still indicate several independent waves of human migration to America, contrary to most recent work on this matter.
Scientists even offer a fairly plausible scenario for the development of these events. The populations that first colonized the northeastern regions of Asia, and subsequently reached and crossed the Bering Strait, and whose descendants eventually populated the territories of South America, were subsequently displaced by a population closer to the modern population of the northeast Asian region and especially the Mongolian ethnic group.
This demonstration of the power of a new statistical approach is sure to stroke the egos of the method's developers, as it can reveal ancient genetic infusion into the hereditary structure of a population, even if the source of this infusion is not preserved today. At the same time, we should not forget that the conclusions of Falus and his colleagues are nothing more than the most plausible scenario. However, almost everything works this way. modern science, trying to recreate the past, inaccessible to our instruments and senses.
Reasons for appearance
genetic differences between populations
People living in different parts of the Earth differ in many ways
characteristics: linguistic affiliation, cultural traditions, appearance,
genetic characteristics. Each population is characterized by its own set
alleles (different states of a gene corresponding to different states
trait, and some alleles may be unique to an ethnic group
or race) and the ratio of their population frequencies.
The genetic characteristics of peoples depend on their history and
lifestyle. In isolated populations that do not exchange gene flow (then
there are no mixes due to geographical, linguistic or religious
barriers), genetic differences arise due to random changes in frequencies
alleles and through the processes of positive and negative natural selection.
Without the influence of any other factors, random changes in genetic
characteristics of populations are usually small.
Significant changes in allele frequencies can occur when
reduction in population size or the resettlement of a small group that provides
the beginning of a new population. Allele frequencies in the new population will be highly dependent
on what the gene pool of the group that founded it was (the so-called founder effect).
The founder effect is associated with an increased frequency of disease-causing mutations in
some ethnic groups.
For example, one type of congenital deafness is caused by
Japanese by a mutation that arose once in the past and is not found in others
regions of the world, that is, all carriers received a mutation from a common ancestor,
which it originated. In white Australians, glaucoma is associated with a mutation
brought by settlers from Europe. A mutation was found in Icelanders,
increasing.the risk of developing cancer and going back to a common ancestor. Similar
the situation was found in residents of the island of Sardinia, but their mutation is different,
different from Icelandic. The founder effect is one of the possible
explanations for the lack of blood type diversity among South American Indians:
their predominant blood group is the first (its frequency is more than 90%, and in many
populations – 100%). Since America was settled by small groups who came
from Asia through the isthmus that once connected these continents, it is possible that in
population that gave rise to the indigenous population of the New World, other blood types
were absent.
Weakly harmful mutations can be maintained in a population for a long time,
whereas mutations that significantly reduce an individual's fitness
are eliminated by selection. It has been shown that disease-causing mutations leading to more
severe forms of hereditary diseases are usually evolutionarily young. For a long time
mutations that have arisen long time persisting in the population are associated with more
mild forms of the disease.
Populations adapt to environmental conditions as a result
selection by fixing randomly occurring new mutations (that is, new
alleles) that increase adaptability to these conditions, and changes in frequencies
existing alleles. Different alleles cause different phenotypes,
for example, skin color or blood cholesterol levels. Allele frequency,
providing an adaptive phenotype (say, dark skin in areas with intense
solar radiation), increases, since its carriers are more viable in data
conditions. Adaptation to different climatic zones manifests itself as variation
frequencies of alleles of a complex of genes, the geographical distribution of which
corresponds to these zones. Most visible footprint in global distribution
genetic variations were left behind by the migration of peoples during their dispersal from the African
ancestral home.
Origin and
human settlement
Earlier history of the appearance of the species Homo sapiens on Earth
reconstructed on the basis of paleontological, archaeological and
anthropological data. In recent decades, the emergence
molecular genetic methods and research of genetic diversity
different peoples made it possible to clarify many questions related to the origin
and the settlement of people of modern anatomical type.
Molecular genetic methods used for
reconstruction of events in demographic history, similar to linguistic ones
methods of proto-language reconstruction. The time that has passed since two
related languages were divided (that is, their common ancestral language ceased to exist
proto-language), assessed by the number of different words that appeared during the period
separate existence of these languages. Similarly, the lifetime of the common
ancestral populations for two modern peoples are estimated by the number
differences (mutations) accumulated in the DNA of representatives of these peoples. Because
the rate of accumulation of mutations in DNA is known by the number of mutations that distinguish two
populations, it is possible to determine when they diverged.
The date of population divergence is determined using:
so-called neutral mutations that do not affect the viability of the individual and do not
subject to the action of natural selection. Such mutations are found in all
regions of the human genome, but most often in phylogenetic studies
consider mutations in DNA contained in cellular organelles - mitochondria
(mtDNA).
First to use mtDNA to reconstruct history
humanity, American geneticist Alan Wilson in 1985. He studied samples
mtDNA obtained from the blood of people from all parts of the world, and based on identified
built a phylogenetic tree of humanity between them. Wilson
showed that all modern mtDNA could have descended from the mtDNA of a common ancestor,
lived in Africa. Wilson's work became widely known. The owner
ancestral mtDNA was immediately dubbed “mitochondrial Eve,” which gave rise to incorrect
interpretations - as if all humanity came from one single woman. On
in fact, “Eve” had several thousand fellow tribesmen, it’s just that their mtDNA was different from ours
the time has not come. However, their contribution is undeniable - we inherited from them
genetic material of chromosomes. The appearance of a new mutation in mtDNA gives rise to
a new genetic line inherited from mother to daughter. Nature of inheritance
in this case can be compared with family property - money and land
can receive from all ancestors, but the surname - from only one of them.
The genetic analogue of the surname transmitted through the female line is mtDNA, through the male line
– Y chromosome, passed from father to son.
To date, the mtDNA of tens of thousands of people has been studied. Managed
isolate mtDNA from the bone remains of ancient people and Neanderthals. Based
studying genetic differences among representatives different nations geneticists have come to
conclusion that over the past million years the number of groups
The number of simultaneously living direct human ancestors ranged from 40 to 100 thousand.
However, about 100-130 thousand years ago, the total number of human ancestors
decreased to 10 thousand individuals (geneticists call the population decline
population with subsequent rapid growth and its passage through the “bottle bottle”
neck"), which led to a significant decrease in genetic diversity
populations (Fig. 1).
Rice. 1. Results of population size assessment based on the study of genetic differences between representatives of different nations.
The reasons for the fluctuation in numbers are still unknown; they are probably
were the same as in other animal species - climate change or food
resources. The described period of population decline and changes in genetic
characteristics of the ancestral population is considered the time of appearance of the species Homo
sapiens.
(Some anthropologists also classify Neanderthals as Homo
sapiens. In this case, the human lineage will be designated as Homo sapiens sapiens, and
Neanderthal - like Homo sapiens neanderthalensis. However, most geneticists
are inclined to believe that Neanderthal represented, although related to man, but
separate species Homo neanderthalensis. These species separated 300-500 thousand years
back.)
mtDNA studies and similar studies of Y chromosome DNA,
transmitted only through the male line, confirmed African origin
people and made it possible to establish the routes and dates of their settlement based on
the spread of various mutations among the peoples of the world. According to modern estimates, the species
Homo sapiens appeared in Africa about 130-180 thousand years ago, then settled in
Asia, Oceania and Europe. America was the last to be populated (Fig. 2).
Rice. 2. Paths (marked by arrows) and dates (indicated by numbers) of human settlement, established on the basis of studying the distribution of various mutations among the peoples of the world.
It is likely that the original ancestral population of Homo sapiens consisted
from small groups living a hunter-gatherer lifestyle. Spreading across
To the earth, people carried with them their traditions and culture and their genes. Perhaps they
also possessed a proto-language. While linguistic reconstructions of the tree
the origin of the world's languages is limited to 30 thousand years, and the existence of a common
of all people of the proto-language is only assumed. And although genes do not determine language,
neither culture, in many cases the genetic kinship of peoples coincides with
the proximity of their languages and cultural traditions. But there are also counter examples,
when peoples changed their language and adopted the traditions of their neighbors. Change of traditions and
language occurred more often in areas of contact of various waves of migration, either as
the result of socio-political changes or conquests.
Of course, in the history of mankind, populations are not only
separated, but also mixed. Therefore, each nation is not represented by only one
genetic line of mtDNA or Y chromosome, but a set of different ones that arose in
different times in different regions of the Earth.
Adaptation of populations
human to living conditions
Results of comparative studies of mtDNA and Y chromosomes
different populations modern people allowed us to hypothesize that
before leaving Africa, about 90 thousand years ago, the ancestral population divided
into several groups, one of which entered Asia through the Arabian Peninsula.
When separated, differences between groups may have been purely due to chance. Big
some racial differences probably arose later as an adaptation to conditions
a habitat. This applies, for example, to skin color - one of the most famous
racial characteristics.
Adaptation to
climatic conditions. The degree of skin pigmentation in humans is genetically
given. Pigmentation provides protection from the damaging effects of the sun
exposure, but should not interfere with receiving the minimum dose
ultraviolet radiation, necessary for the formation of vitamin D in the human body,
preventing rickets.
In northern latitudes, where radiation intensity is low, people
have lighter skin. Residents of the equatorial zone have the darkest
skin. The exceptions are the inhabitants of shaded tropical forests - their skin
lighter than would be expected for these latitudes, and some northern peoples
(Chukchi, Eskimos), whose skin is relatively highly pigmented, since they
eat foods rich in vitamin D, such as marine liver
animals. Thus, differences in the intensity of ultraviolet radiation
act as a selection factor, leading to geographical variations in skin color.
Light skin is an evolutionarily later trait that arose due to mutations in
several genes that regulate the production of the skin pigment melanin. Ability
Sunbathing is also determined genetically. It is distinguished by residents of regions with
strong seasonal fluctuations in solar radiation intensity.
There are known climatic differences in
body structure. We are talking about adaptations to cold or warm climates:
short limbs in Arctic populations (Chukchi, Eskimos) increase
the ratio of body mass to its surface and thereby reduce heat transfer, and
inhabitants of hot, dry regions, such as the African Maasai, are distinguished by long
limbs. Inhabitants of areas with a humid climate are characterized by wide and
flat noses, and in dry cold climates a long nose is more effective, better
warming and moisturizing inhaled air.
Adaptation to life in high mountain conditions is
increased hemoglobin content in the blood and increased pulmonary blood flow. Such
features are observed among the indigenous inhabitants of the Pamirs, Tibet and the Andes. All these
differences are determined genetically, but the degree of their manifestation depends on the conditions
development in childhood. For example, among Andean Indians who grew up at sea level,
signs are less pronounced.
Adaptation to types
nutrition. Some genetic changes are associated with differences in types
nutrition. The most famous among them is hypolactasia - milk intolerance.
sugar (lactose). To digest lactose, young mammals produce
lactase enzyme. At the end of the feeding period, this enzyme disappears from
intestinal tract of the cub and in adults is not produced.
The absence of lactase in adults is initial, ancestral
sign for a person. In many Asian and African countries where adults
traditionally do not drink milk; after the age of five, lactase stops
be developed. Drinking milk under such conditions leads to disorder
digestion. However, most European adults produce lactase and
can drink milk without harm to health. These people are carriers of the mutation
in the DNA region that regulates lactase synthesis. The mutation spread after
the emergence of dairy farming 9-10 thousand years ago and occurs
mainly among European peoples. More than 90% of Swedes and Danes are able
digest milk, and only a small part of the Scandinavian population differs
hypolactasia. In Russia, the incidence of hypolactasia is about 30% for Russians and
more than 60-80% for the indigenous peoples of Siberia and the Far East.
Peoples in whom hypolactasia is combined with breast milk
cattle breeding, traditionally they eat not raw milk, but fermented milk
products in which milk sugar has already been processed by bacteria into easily
digestible substances. The predominance of a one-size-fits-all Western-style diet in
in some countries leads to the fact that some children with undiagnosed
hypolactasia reacts to milk with indigestion, which is taken
for intestinal infections. Instead of the diet change necessary in such cases
antibiotic treatment is prescribed, leading to the development of dysbacteriosis. More
one factor could contribute to the spread of lactase synthesis in adults - in
In the presence of lactase, milk sugar promotes the absorption of calcium, performing those
the same functions as vitamin D. Perhaps this is why northern Europeans
mutation about which we're talking about, occurs most often.
Residents of North Asia are distinguished by a hereditary lack of
trehalase enzyme, which breaks down mushroom carbohydrates, which are traditionally
They are considered here as food for deer, not suitable for humans.
The population of East Asia is characterized by a different
hereditary feature of metabolism: many Mongoloids even from small
doses of alcohol quickly make you drunk and can cause severe intoxication due to
accumulation of acetaldehyde in the blood, formed during the oxidation of alcohol
liver enzymes. Oxidation occurs in two stages: in the first, ethyl alcohol
turns into toxic ethyl aldehyde, on the second the aldehyde is oxidized with
the formation of harmless products that are excreted from the body. Speed
work of enzymes of the first and second stages (with unreadable names
alcohol dehydrogenase and acetal dehydrogenase) are genetically determined.
In East Asia, the combination of “fast” is common
enzymes of the first stage with “slow” enzymes of the second, that is, when taking
alcohol, ethanol is quickly processed into aldehyde (first stage), and its
further removal (second stage) occurs slowly. This feature
Eastern Mongoloids is due to the frequent combination of two mutations in them,
affecting the speed of operation of the mentioned enzymes. It is supposed to be so
adaptation to a still unknown environmental factor is manifested.
Adaptations to the type of nutrition are associated with complexes of genetic
changes, few of which have yet been studied in detail at the DNA level. For example, about
20-30% of the inhabitants of Ethiopia and Saudi Arabia are able to quickly break down some
nutritional substances and drugs, in particular amitriptyline, due to the presence of
two or more copies of a gene encoding one of the types of cytochromes -
enzymes that break down foreign substances that enter the body with food. U
peoples of other regions, doubling of this gene occurs with a frequency of no more than
3-5%. It is believed that the increase in the number of gene copies is caused by diet
(possibly by eating large quantities of pepper or edible plant
teff, which makes up up to 60% of food in Ethiopia and nowhere else
widespread to such an extent). But what is the cause and what is the effect?
impossible to determine at present. Did the accidental.increase
frequencies in a population of carriers of multiple genes to the fact that people were able to eat
any special plants? Or that they started eating pepper (or
any other product that requires this cytochrome for absorption)
caused an increase in the frequency of gene doubling? Either of these two processes could
take place during the evolution of populations.
It is obvious that the food traditions of the people and genetic factors
interact. Consumption of certain types of food becomes possible only
in the presence of certain genetic prerequisites, and which subsequently became
traditional diet acts as a selection factor and leads to changes in frequencies
alleles and distribution of genetic variants in the population, the most
adaptive for this diet. Traditions usually change slowly. So, the transition from
gathering to agriculture and the accompanying changes in diet and lifestyle
lives continued for tens and hundreds of generations. Relatively slow
Changes in the gene pool of populations that accompany such events also occur.
Allele frequencies can change by 2-5% per generation, and these changes
accumulate from generation to generation. The action of other factors, for example
epidemics, often associated with wars and social crises, can be several
change allele frequencies once during the life of one generation due to
a sharp decline in population size. So, the conquest of America by Europeans
led to the death of 90% of the indigenous population as a result of wars and epidemics.
Genetics of resistance
to infectious diseases
Sedentary lifestyle, development of agriculture and cattle breeding,
increased population density contributed to the spread of infections and
outbreaks of epidemics. For example, tuberculosis is formerly a disease of cattle
livestock, was obtained by humans after the domestication of animals and became epidemic
significant in the emergence and growth of cities. Epidemics have made the problem urgent
resistance to infections. Resistance to infections is also genetic
component.
The first example of sustainability studied is
spread of hereditary disease in tropical and subtropical zones
blood - sickle cell anemia, which is caused by a mutation in the gene
hemoglobin, leading to disruption of its functions. In patients, the shape of red blood cells,
determined by a microscopic blood test, not oval, but crescent-shaped,
This is where the disease got its name. The carriers of the mutation turned out to be
resistant to malaria. In areas where malaria is widespread, it is most “profitable”
heterozygous state (when from a pair of genes obtained from
parents, only one is damaged, the other is normal), since homozygous
carriers of mutant hemoglobin die from anemia, homozygous for normal
gene - suffer from malaria, and in heterozygous anemia manifests itself in a mild form and
they are protected from malaria.
Another hereditary disease is common in Europe -
cystic fibrosis. Its cause is a mutation that disrupts the regulation of salt metabolism and
water balance of cells. In patients, all organs secreting mucous membranes are affected
secretions (bronchopulmonary system, liver, various glands). They die by
adolescence, leaving no offspring. However, the disease occurs
only if the child receives a damaged gene from both parents,
heterozygous mutation carriers are quite viable, although the release of glandular
their secretions and fluid levels may be reduced.
In Europe, cystic fibrosis affects one in 2,500
born. In the heterozygous state, the mutation is present in one in 50
humans – a very high frequency for a pathogenic mutation. Therefore it should
assume that natural selection acts in favor of its accumulation in
populations, that is, heterozygotes have increased fitness. AND
indeed, they are believed to be more resistant to intestinal infections.
There are several hypotheses about the mechanisms of this resistance. According to one of
Among them, heterozygotes for the mutation have reduced fluid secretion through the intestines, so
that they are less likely to die from dehydration due to diarrhea that occurs
as a result of infection. But in hot climates, harm from salt imbalance
exchange outweighs the benefits of increased resistance to infection - and
Cystic fibrosis is extremely rare there due to reduced vitality
carriers of mutations.
Resistance to tuberculosis is associated with spread in
some populations of Tay-Sachs disease, a severe hereditary disease,
leading to degeneration nervous system and changes in the respiratory mucosa
tract. A gene has been identified whose mutations lead to the development of the disease.
It is assumed that heterozygous mutation carriers are more resistant to tuberculosis.
These examples show that the population pays for increasing
the survival rate of heterozygous mutation carriers may be an order of magnitude higher
less common homozygous carriers, which inevitably appear when
increasing its population frequency. However, mutations are known that are also
homozygous state protects against infections, such as viral infection
human immunodeficiency, HIV, or slow down the development of the disease after
infection. Two such mutations occur in all populations, and another
of European origin, and is absent in other regions. Supposed,
that these mutations spread in the past because they are protective
effect on other epidemic diseases. In particular,
the spread of the mutation among Europeans is associated with the “Black Death” epidemic
plague, which in the 14th century wiped out a third of the population of Europe, and in some regions - up to
80%. Another candidate for the role of a selection factor is smallpox, which also carried away many
lives. Before the emergence of large cities and reaching the epidemic threshold
population size, such large-scale “selection rounds” for resistance to
infections were impossible.
Development of civilization and
genetic changes
It seems surprising that the Bushmen's diet is
hunter-gatherers living in South Africa turned out to be appropriate
WHO recommendations on the overall balance of proteins, fats, carbohydrates, vitamins,
microelements and calories. Biologically, man and his immediate ancestors are
over hundreds of thousands of years, they adapted to the hunter-gatherer lifestyle.
Changing traditional diet and lifestyle
affects people's health. For example, African Americans are more likely than Euro-Americans
suffer from hypertension. North Asian peoples, whose traditional diet was
rich in fats, the transition to European high-carbohydrate foods leads to the development
diabetes and other diseases.
The previously prevailing ideas that with the development
productive economy (farming and livestock breeding) health and nutrition of people
steadily improving, now refuted: many common diseases
were rarely found among ancient hunter-gatherers, if at all
unknown. With the transition to agriculture, life expectancy decreased (from
30-40 years to 20-30), the birth rate increased by 2-3 and at the same time significantly
child mortality increased. Bony remains of early agricultural peoples
more often have signs of previous anemia, malnutrition, and various infections than
pre-agricultural.
Only in the Middle Ages did a turning point come - and the duration
life began to increase. Significant improvement in population health in developed countries
countries is associated with the advent of modern medicine.
To the factors that distinguish modern agricultural peoples,
include a high-carbohydrate and high-cholesterol diet, salt intake, decreased
physical activity, sedentary lifestyle, high population density,
complication of social structure. Adaptation of populations to each of these factors
accompanied by genetic changes, that is, an increase in the frequency
adaptive alleles in the population. The frequency of non-adaptive alleles decreases,
because their carriers are less viable or have fewer numbers
descendants. Thus, the low-cholesterol diet of hunter-gatherers makes
adaptive for them is the ability to intensively absorb cholesterol from food,
what about modern look life becomes a risk factor for atherosclerosis and
cardiovascular diseases. Efficient salt absorption, beneficial in the past,
when salt was unavailable, becomes a risk factor for hypertension. Changes
population frequencies of alleles during man-made transformation of the habitat
human developments occur in the same way as during adaptation to natural conditions. Recommendations
health care doctors ( physical activity, taking vitamins and
microelements, salt restriction) artificially recreate the conditions in which
man lived most of the time of his existence as a biological species.
Ethical considerations
studying genetic differences between people
So, the formation of gene pools of ethnic groups is influenced by
various processes - accumulation of mutations in isolated groups, migration and
mixing of peoples, adaptation of populations to environmental conditions. Genetic differences
do not imply the superiority of any race, ethnicity or education
any other characteristic (type of economy or level of complexity of social
organizations) groups. On the contrary, they emphasize the evolutionary value
diversity of humanity, which allowed it to populate all climatic zones
Earth.
Magazine "Energy" 2005, No. 8
scientific co-workers Laboratory of Genome Analysis, Institute of General Genetics
them. N.I. Vavilova RAS
Genetic diversity of peoples
 People living in different parts of the Earth differ in many ways: language, cultural traditions, appearance, genetic characteristics.
People living in different parts of the Earth differ in many ways: language, cultural traditions, appearance, genetic characteristics.
The genetic characteristics of peoples depend on their history and lifestyle. Differences between them arise in isolated populations that do not exchange gene flow (i.e., do not mix due to geographic, linguistic, or religious barriers), through random changes in allele frequencies and processes of positive and negative natural selection. A random change in allele frequencies in a population is called genetic drift . The differences in these frequencies without the influence of any additional factors are usually small. When a population declines or a small group emigrates to give rise to a new population, allele frequencies can fluctuate greatly. In the new population they will depend on the gene pool of the group that founded it (the so-called
Weakly harmful mutations can be maintained in a population for a long time, while harmful ones, which significantly reduce the fitness of an individual, are eliminated by selection. It has been shown that pathogenic mutations that cause severe forms of hereditary diseases are usually evolutionarily young. Long-established mutations that persist for a long time in the population are associated with milder forms of the disease.
Adaptation to environmental conditions is fixed during selection due to randomly arising new alleles that increase fitness to given conditions, or due to changes in the frequencies of long-existing alleles.
Different alleles cause variations in phenotype, such as skin color or blood cholesterol levels. The frequency of an allele that provides an adaptive phenotype (for example, dark skin in areas with intense solar radiation) increases because its carriers are more viable in these conditions.
Adaptation to different climatic zones manifests itself as a variation in the allele frequencies of a complex of genes, the geographical distribution of which corresponds to the climatic zones.
However, the most noticeable mark on the global distribution of genetic changes was left by migrations of peoples associated with resettlement from the African ancestral home. Origin and settlement of man Previous history of the appearance of the species
Homo sapiens
To date this event, neutral mutations are used that do not affect the viability of the individual and are not subject to the action of natural selection. They are found in all parts of the human genome, but most often they use mutations in the DNA contained in cellular organelles - mitochondria. In a fertilized egg, only maternal mitochondrial DNA (mtDNA) is present, since the sperm does not transfer its mitochondria to the egg. For phylogenetic studies, mtDNA has particular advantages. Firstly, it does not undergo recombination like autosomal genes, which greatly simplifies the analysis of pedigrees.
Secondly, it is contained in a cell in quantities of several hundred copies and is much better preserved in biological samples.
The first to use mtDNA to reconstruct human history was American geneticist Alan Wilson in 1985. He studied mtDNA samples obtained from the blood of people from all parts of the world, and based on the differences identified between them, he built a phylogenetic tree of humanity. It turned out that all modern mtDNA could have come from the mtDNA of a common ancestor who lived in Africa. The owner of the ancestral mtDNA was immediately dubbed “mitochondrial Eve,” which gave rise to incorrect interpretations - that all of humanity descended from one single woman. In fact, “Eve” had several thousand fellow tribesmen, but their mtDNA has not reached our times. However, they all undoubtedly left their mark: from them we inherited the genetic material of the chromosomes. The nature of inheritance in this case can be compared to family property: a person can receive money and land from all ancestors, but a surname - from only one of them. The genetic analogue of a surname transmitted through the female line is mtDNA, and in the male line it is the Y chromosome, transmitted from father to son. The study of mtDNA and Y-chromosome DNA confirmed the African origin of man and made it possible to establish the routes and dates of his migration based on the distribution of various mutations among the peoples of the world. According to modern estimates, the species
H.sapiens The nature of inheritance in this case can be compared to family property: a person can receive money and land from all ancestors, but a surname - from only one of them. The genetic analogue of a surname transmitted through the female line is mtDNA, and in the male line it is the Y chromosome, transmitted from father to son. consisted of small groups leading the life of hunter-gatherers. When migrating, people carried with them their traditions, culture and their genes. Perhaps they also possessed a proto-language.
So far, linguistic reconstructions of the origin of the world's languages are limited to a period of 15–30 thousand years, and the existence of a common proto-language is only assumed. And although genes do not determine either language or culture, in some cases the genetic relatedness of peoples also coincides with the similarity of their languages and cultural traditions. But there are also opposite examples, when peoples changed their language and adopted the traditions of their neighbors.
Such a change occurred more often in areas of contact between different waves of migration or as a result of socio-political changes or conquests.
Climate. One of the most well-known racial characteristics is skin color, the pigmentation of which in humans is genetically determined.

Pigmentation protects against the damaging effects of solar radiation, but should not interfere with receiving the minimum dose of radiation necessary to produce vitamin D, which prevents rickets. In northern latitudes, where radiation intensity is low, people have lighter skin, and in the equatorial zone it is the darkest. However, the inhabitants of shaded tropical forests have lighter skin than would be expected at a given latitude, and some northern peoples (Chukchi, Eskimos), on the contrary, have relatively highly pigmented skin. In the latter case, this is explained either by the supply of vitamin D with food (fish and liver of marine animals), or by the recent migration of northern groups from lower latitudes on an evolutionary scale.
Thus, the intensity of ultraviolet radiation acts as a selection factor, leading to geographic variations in skin color. Light skin is an evolutionarily later trait and arose due to mutations in several genes that regulate the production of the skin pigment melanin (melaninocortin receptor gene MC1R and others).
The ability to tan is also genetically determined. It is distinguished by residents of regions with strong seasonal fluctuations in the intensity of solar radiation.
Differences in body structure associated with climatic conditions are known. These are adaptations to cold or warm climates. Thus, short limbs of residents of the Arctic regions (Chukchi, Eskimos) reduce the ratio of body surface to its mass and thereby reduce heat transfer. Inhabitants of hot, dry regions, such as the African Masai, on the contrary, are distinguished by long limbs. Those living in humid climates have wider, flatter noses, while those in dry, colder climates have longer noses, which helps warm and moisten the air they inhale. hypolactasia. The young of all mammals produce the enzyme lactase to digest lactose. At the end of feeding, it disappears from the intestinal tract of the calf. The absence of the enzyme in adults is the initial, ancestral trait for humans.
In many Asian and African countries where adults traditionally do not drink milk, lactase is not synthesized after the age of five, and therefore drinking milk leads to indigestion. However, most adult Europeans can drink milk without harm to their health: their lactase synthesis does not stop due to a mutation in the DNA section that regulates the formation of the enzyme. This mutation spread after the advent of dairy farming 9–10 thousand years ago and is found mainly among European peoples. More than 90% of Swedes and Danes are able to digest milk, and only a small part of the Scandinavian population is hypolactasic. In Russia, the incidence of hypolactasia is about 30% for Russians and more than 60–80% for indigenous peoples of Siberia and the Far East. Peoples whose hypolactasia is combined with dairy farming traditionally use not raw milk, but fermented milk products in which milk sugar has already been broken down by bacteria.
The lack of information about the genetic characteristics of people sometimes leads to the fact that with hypolactasia, people who react to milk with indigestion, which is mistaken for intestinal infections, are prescribed antibiotic treatment, leading to dysbiosis, instead of the necessary diet change.
In addition to milk consumption, another factor could influence the persistence of lactase synthesis in adults. In the presence of lactase, milk sugar promotes the absorption of calcium, performing the same functions as vitamin D. This may be why the mutation in question is most common in northern Europeans. This is an example of genetic adaptation to interacting dietary and climatic factors.
A few more examples. Eskimos with a traditional diet usually consume up to 2 kg of meat per day. Digesting such quantities of meat is possible only with a combination of certain cultural (culinary) traditions, a certain type of microflora and hereditary physiological characteristics of digestion.
Among the peoples of Europe it occurs celiac disease– intolerance to the gluten protein contained in rye, wheat and other grains. When consuming cereals, it causes multiple developmental disorders and mental retardation. The disease is 10 times more common in Ireland than in continental Europe, probably because wheat and other grains were not traditionally staple foods there.
Residents of the North Asian region often lack the enzyme trehalase, which breaks down mushroom carbohydrates. This hereditary feature is combined with a cultural one: in these places, mushrooms are considered food for deer, not suitable for humans.

Residents of East Asia are characterized by another hereditary metabolic feature. It is known that many Mongoloids quickly become drunk even from small doses of alcohol and can become severely intoxicated. This is due to the accumulation of acetaldehyde in the blood, which is formed during the oxidation of alcohol by liver enzymes. It is known that alcohol is oxidized in the liver in two stages: first it turns into toxic acetaldehyde, and then it is oxidized to form harmless products that are excreted from the body. The speed of the enzymes of the first and second stages (alcohol dehydrogenase and acetal dehydrogenase) is determined genetically. The indigenous population of East Asia is characterized by a combination of “fast” enzymes of the first stage with “slow” enzymes of the second stage. In this case, when drinking alcohol, ethanol is quickly processed into aldehyde (first stage), and its further removal (second stage) occurs slowly. This feature is associated with a combination of two mutations that affect the speed of operation of these enzymes. It is assumed that the high frequency of these mutations (30–70%) is the result of adaptation to an as yet unknown environmental factor.
Adaptations to the type of nutrition are associated with complexes of genetic changes, not many of which have yet been studied in detail at the DNA level.
It is known that about 20-30% of residents of Ethiopia and Saudi Arabia are able to quickly break down certain nutrients and drugs, in particular amitriptyline, due to the presence of two or more copies of the gene encoding one of the types of cytochromes - enzymes that decompose foreign substances entering the body with food. In other peoples, doubling of this cytochrome gene occurs with a frequency of no more than 3–5%, and inactive variants of the gene are common (from 2–7% in Europeans and up to 30% in China). It is possible that the copy number of the gene increases due to dietary patterns (the use of large quantities of pepper or the edible plant teff, which makes up up to 60% of the food supply in Ethiopia and is not common to such an extent anywhere else). However, it is currently impossible to determine where the cause is and where the effect is. Is it a coincidence that an increase in the population of carriers of multiple genes allowed people to eat some special plants? Or, conversely, did the consumption of pepper (or other foods that require cytochrome for absorption) serve as a factor in the selection of individuals with a doubled gene? Both one and the other process could have taken place in the evolution of populations.
It is obvious that the food traditions of a people and genetic factors interact.
Eating a particular food becomes possible only if certain genetic prerequisites are present, and a diet that has become traditional acts as a selection factor, influencing the frequency of alleles and the distribution in the population of the genetic variants that are most adaptive for such nutrition.
A sedentary lifestyle, the development of agriculture and cattle breeding, and an increase in population density contributed to the spread of infections and the emergence of epidemics. Thus, tuberculosis, originally a disease of cattle, was acquired by humans after the domestication of animals. With the growth of cities, the disease has become epidemically significant, which has made resistance to the infection, which also has a genetic component, relevant.
The most thoroughly studied example of such resistance is the spread of the disease sickle cell anemia in tropical and subtropical zones, so named because of the sickle-shaped shape of the red blood cells (determined by microscopic analysis of a blood smear). This hereditary disease is caused by a mutation in the hemoglobin gene, leading to disruption of its functions. Carriers of the mutation turned out to be resistant to malaria. In areas where the disease is widespread, the heterozygous state is most adaptive: homozygotes with mutant hemoglobin die from anemia, homozygotes with a normal gene suffer from malaria, and heterozygotes in whom anemia manifests itself in a mild form are protected from malaria.
Such examples show that the price for the increased adaptability of heterozygotes can be the death of an order of magnitude less common homozygotes for a pathogenic mutation, which inevitably appear with an increase in its population frequency.
Another example of genetic determination of susceptibility to infections is the so-called prion diseases. These include spongiform disease of the brain of cattle (mad cow disease), an outbreak of which among cattle was observed after the advent of a new technology for processing bone meal used for animal feed. The infection is transmitted to humans with very low frequency through the meat of sick animals. The few people who became ill turned out to be carriers of a rare mutation, previously considered neutral.
There are mutations that protect against infection with the human immunodeficiency virus or slow down the progression of the disease after infection. Two of these mutations are found in all populations (with a frequency of 0 to 70%), and another is found only in Europe (frequency of 5–18%). These mutations are thought to have spread in the past because they have a protective effect 2 against other epidemic diseases.
Development of civilization and genetic changes
It seems surprising that the diet of the Bushmen - hunter-gatherers living in South Africa - turned out to be fully consistent with WHO recommendations for the overall balance of proteins, fats, carbohydrates, vitamins, microelements and calories. Biologically, humans and their immediate ancestors adapted to the hunter-gatherer lifestyle over hundreds of thousands of years.
Changes in traditional nutrition and lifestyle affect people's health.
For example, African Americans are more likely than Euro-Americans to have hypertension. Among northern peoples, whose traditional diet was rich in fat, the transition to a European high-carbohydrate diet contributes to the development of diabetes and other diseases. The previously prevailing ideas that with the development of the productive economy (farming and cattle breeding) people's health and nutrition are steadily improving have now been refuted. After the advent of agriculture and animal husbandry, many diseases that were rare or completely unknown to ancient hunter-gatherers became widespread.
Today, agricultural peoples are characterized by a high-carbohydrate and high-cholesterol diet, the use of salt, decreased physical activity, a sedentary lifestyle, high population density, and a more complex social structure. The adaptation of populations to each of these factors is accompanied by genetic changes: there are more adaptive alleles, and fewer non-adaptive alleles, since their carriers are less viable or less fertile. For example, the low-cholesterol diet of hunter-gatherers makes them adaptive for the ability to intensively absorb cholesterol from food, but with modern lifestyles it becomes a risk factor for atherosclerosis and cardiovascular diseases. Effective absorption of salt, which was useful when it was unavailable, in modern conditions turns into a risk factor for hypertension.
During man-made transformation of the human environment, population allele frequencies change in the same way as during natural adaptation.
Doctors' recommendations for maintaining health - physical activity, taking vitamins and microelements, limiting salt, etc. – in fact, they artificially recreate the conditions in which man lived for most of his existence as a biological species. It is likely that certain adaptations could be associated with the collective way of life of humans. Thus, the increased incidence of depression in modern societies Western type is caused by the loss of support from the clan group. A number of studies have shown that with the destruction of the birth system, the survival rate of children decreases and the risk of developing diseases increases. According to statistics, the incidence of depression varies significantly in different countries
So, the formation of gene pools of ethnic groups is influenced by many processes: migration and mixing of peoples, accumulation of mutations in isolated groups, adaptation of populations to environmental conditions. Interpopulation (geographical, linguistic and other) barriers contribute to the accumulation of genetic differences, which, however, are usually not very significant between neighbors. The geographic distribution of these differences reflects a continuum of changing traits and changing gene pools. Genetic differences do not imply the superiority of any race, ethnic or other group formed on any basis (type of economy or social organization). On the contrary, they emphasize the evolutionary value of diversity, which allowed humanity not only to master all the climatic zones of the Earth, but also to adapt to those significant environmental changes that arose as a result of human activity.
Literature
Gene pool and genogeography of the population of Russia and neighboring countries / Ed. SOUTH. Rychkova. – St. Petersburg, 2000.
Gorbunova V.N., Baranov V.S. Introduction to molecular diagnostics and gene therapy of hereditary diseases. – St. Petersburg, 1997.
Limborskaya S.A., Khusnutdinova E.K., Balanovskaya E.V. Ethnogenomics and genogeography of peoples of Eastern Europe. – M., 2002.
Stepanov V.A. Ethnogenomics of the peoples of Northern Eurasia - Tomsk, 2002.
Evolution in health and disease / Ed. S.C. Stearns. – N.Y., 1999.
Cavalli-Sforza L.L., Menozzi P., Piazza A. History and Geography of Human Genes, Princeton. – N.Y., 1994.
Cavalli-Sforza L.L. Genes, peoples, languages // In the world of science. 1992.
Wilson A.K., Kann R.L. Recent African origin of people // In the world of science.
1992. Borinskaya S.A., Khusnutdinova E.K.
Ethnogenomics: history with geography // Man. 2002. No. 1. P. 19–30. Khusnutdinova E.K., Borinskaya S.A.
Genomic medicine – medicine of the 21st century // Nature. 2002. No. 12. P.3–8.
Human genome: threads of fate // Chemistry and life. 1998. No. 4. P.27–30. Yankovsky N.K., Borinskaya S.A.
Our history recorded in DNA // Nature. 2001. No. 6. P.10–17.